Motivation¶
What’s in a domain?¶
The first large-scale three-dimensional features about chromatin gleaned from early Hi-C studies were the open and closed regions of chromatin called compartments (see Figure ). More recent Hi-C mapping identified high contact density regions within chromatin compartments called topologically associating domains (TADs) that are largely conserved in their positions between cell types . The exact boundaries were determined by training a hidden Markov model against computed tracks of contact frequency directionality bias. However, as can be seen by eye in Figure , such regions can be further stratified into regions both larger and smaller than the ones defined as TADs (green line segments). This makes it challenging to determine the biological significance of domains at any particular scale and highlights the importance of assessing structural signatures at many different scales and resolutions in order to identify more precisely what features are predictive of biological function.
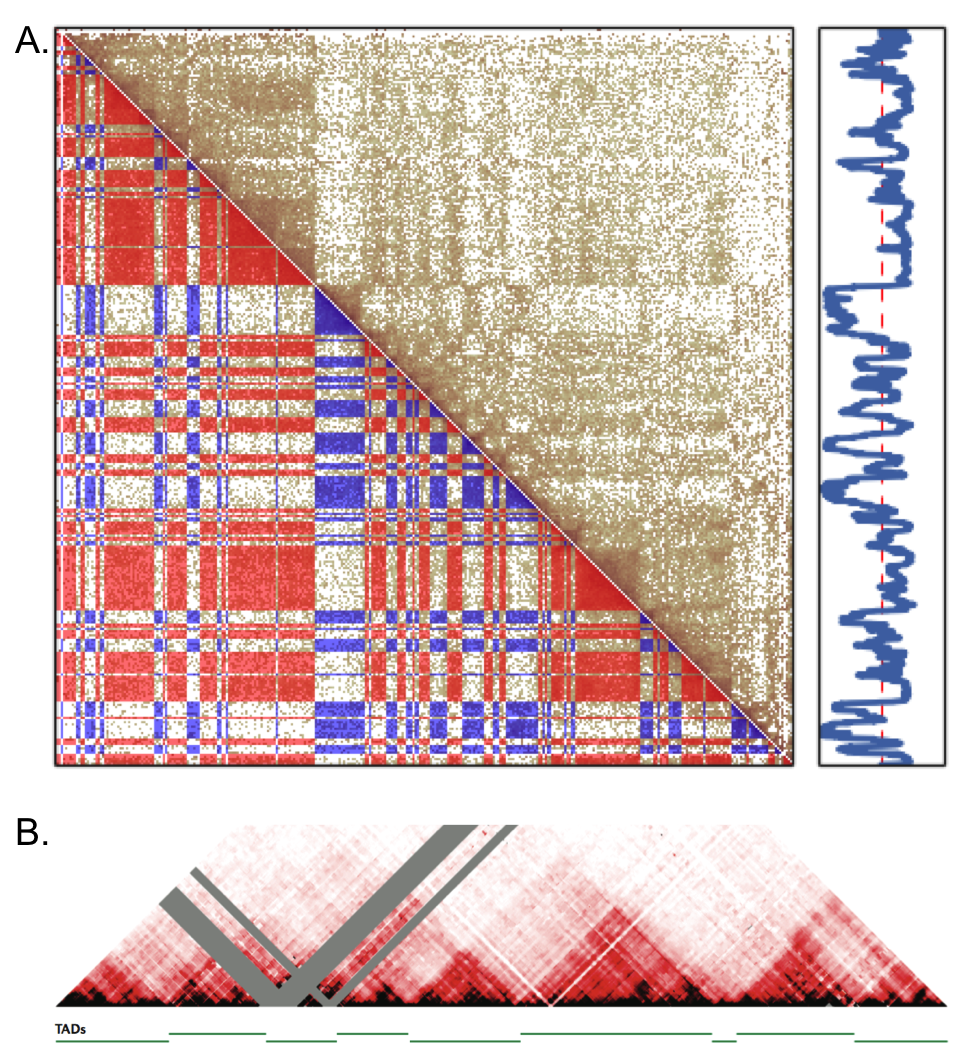
Fig. 1
Finding communities in a haystack¶
Hi-C heatmaps \(A_{ij}\) provide quantitative readouts for pairs of genomic coordinate bins \(i\) and \(j\). We can regard a symmetric heatmap matrix \(A\) as the adjacency matrix of an undirected weighted graph, whose nodes are genomic bins (see Fig. 1). The edge weight \(A_{ij}\) is an interaction score for bins \(i\) and \(j\). In the case of processed Hi-C data, this is normally interpreted as the relative contact frequency between the two genomic loci.
A network-oriented perspective makes it tempting to apply graph algorithms for node partitioning, also known as community detection in the field of network science, to identify biologically relevant domains. One of the most popular methods for community detection in networks is modularity maximization . For a given assignment of nodes to communities (i.e., partition), an objective function called modularity compares the edge weight between pairs of nodes in the same community to the expected edge weight between two nodes under a random null model. The null model most commonly used is called the configuration null model, which consists of the complete ensemble of graphs having a fixed degree distribution. Under the configuration null model, the expected weight between two nodes \(i\) and \(j\) of degree \(k_i\) and \(k_j\) is \(\frac{k_i k_j}{2m}\), where \(m\) is the total edge weight of the graph.
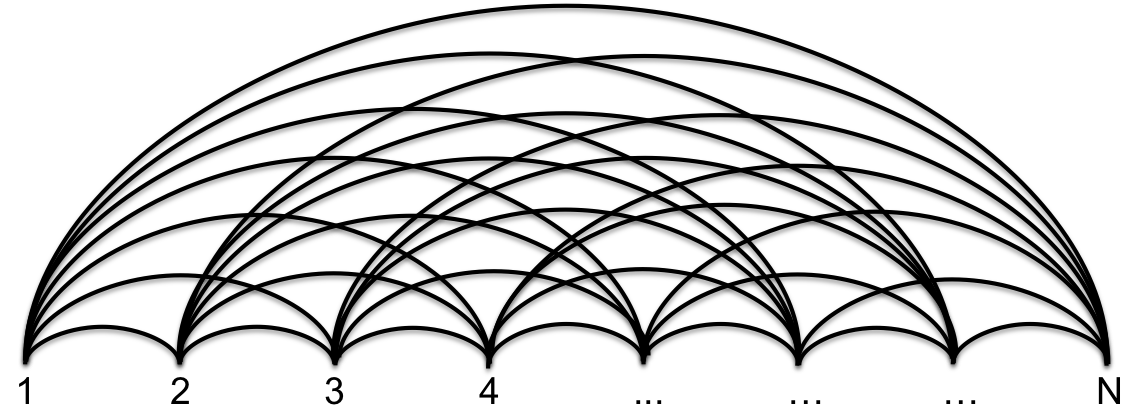
Fig. 2
The multi-resolution form of the modularity function \(Q\) for a partition \(\pi\) of a network’s nodes \(\{1,2, \ldots, N\}\) is
where the summation runs over all pairs of nodes in the same community (\(\delta(\pi_i, \pi_j) = 1\) if \(\pi_i=\pi_j\) and \(0\) if not). The parameter \(\gamma\) acts as a weight on the null model, and determines the resolution of the partition. The larger the value of \(\gamma\), the smaller the typical size of detected communities.
Models of interacting spin systems from statistical physics have played a key role in the development of modern algorithms for community detection. The most well known spin system is the Ising model, which is a model of ferromagnetism, where magnetic spins are nodes on a lattice that take on one of two states (up or down) and interact with their nearest neighbors. Modularity maximization is actually equivalent to finding the ground state of a particular kind of spin system known as the Potts model where each node in the network corresponds to a magnetic spin that can assume one of a number of different spin states (i.e. the community labels). Edge weights between nodes in the same spin state (community) are ferromagnetic (stabilizing) coupling energies. The null model acts as an anti-ferromagnetic (destabilizing) applied field on interacting spins. Finding the maximum modularity partition of the graph therefore corresponds to minimizing the Potts Hamiltonian \(H_{potts}(\pi) = -mQ(\pi)\) to find the ground state configuration of the spins.
Restrict the search space¶
In general, modularity maximization is NP-complete cite{}, but we gain a great deal of traction if we restrict the solution space of partitions of the nodes to segmentations, where only chains of consecutive nodes can be grouped together. This is convenient because many of the interesting features seen by eye in Hi-C data, such as TADs and their apparent substrata, are segmental. Not only can we compute a single point estimate of the community structure from the data, but we can efficiently extract a great deal of information about entire statistical ensembles of segmentations by dynamic programming, and thereby characterize features at multiple scales.